Highlights
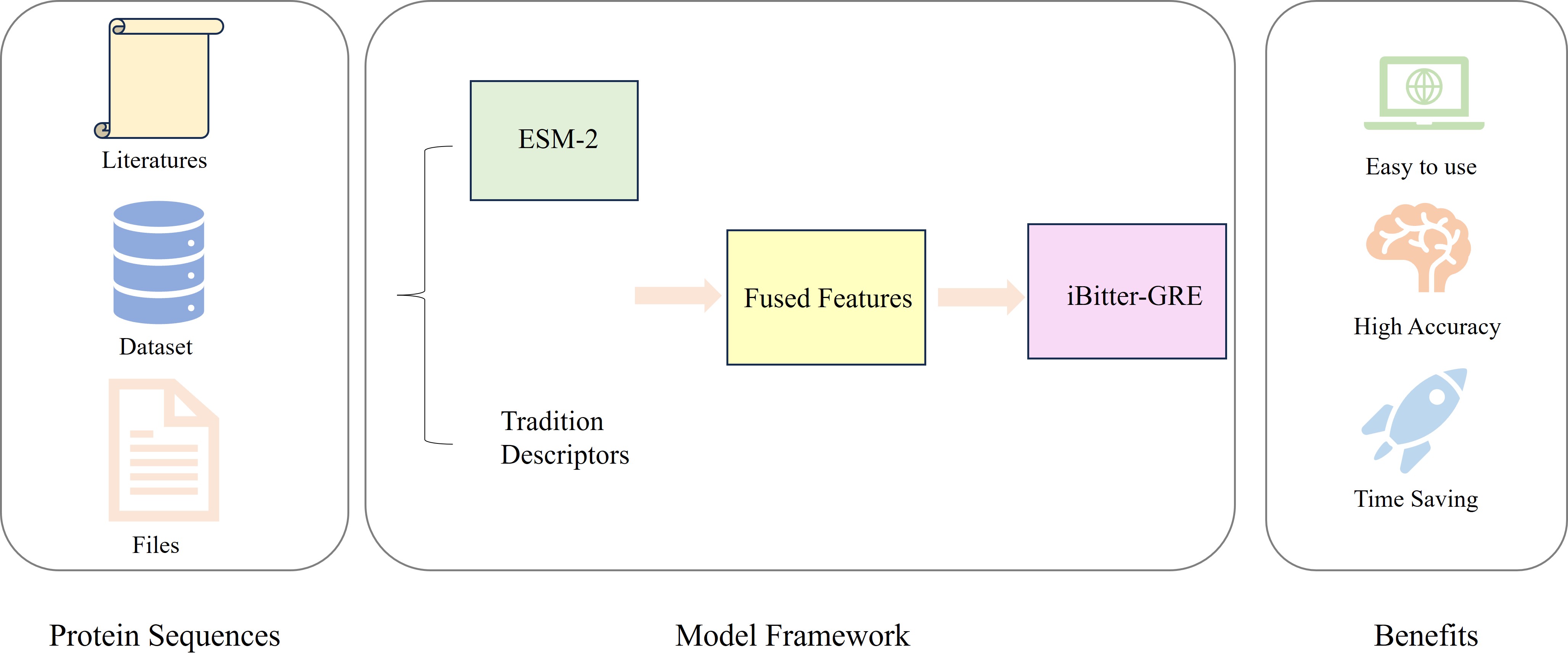
- In this study, we present iBitter-GRE, a powerful bioinformatics tool for identifying bitter peptides.
- iBitter-GRE combines ESM-2 and traditional features to learn structure-related and physicochemical information.
- iBitter-GRE integrates three models: GB, RF, and ETree, ensuring the robustness of the model.
- SHAP is applied to explain the features extracted by traditional methods.
- Comparative studies show that iBitter-GRE outperforms several existing predictors. The tool is publicly accessible.