Publications
Our current and previous research highlights.
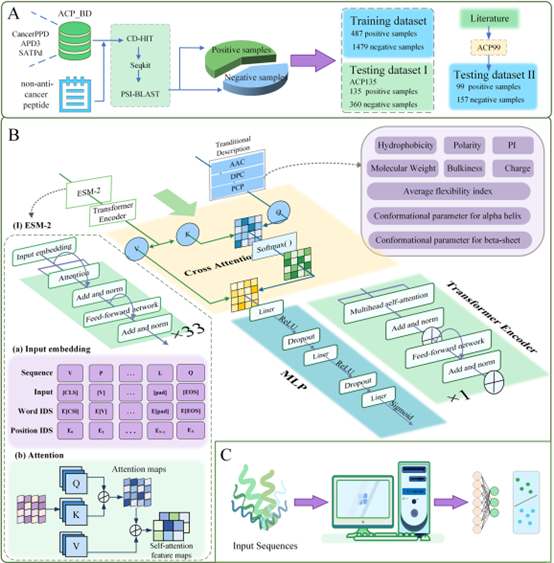
ACP ACP-EPC: An Interpretable Deep Learning Framework for Anticancer Peptide Prediction Utilizing Pre-trained Protein Language Model and Multi-view Feature Extracting Strategy
Background and Objective
Cancer remains a significant global health challenge, and traditional chemotherapy frequently damages healthy cells, leading to severe side effects. Anticancer peptides (ACPs) present a promising alternative as they specifically target and eliminate cancer cells, enhancing treatment outcomes and patient quality of life. However, traditional biological methods for identifying ACPs are complex and time-consuming. Thus, this study aims to develop an efficient and accurate computational method for predicting ACPs.
Methods
We developed a deep learning model designed to predict anticancer peptides based on protein sequences. Our model uniquely integrates ESM-2 for structural and evolutionary feature extraction with traditional physicochemical feature extraction methods. Additionally, our model employs a cross-attention mechanism to effectively fuse multimodal features, enhancing robustness and interpretability.
Results
Our model was rigorously evaluated using 10-fold cross-validation and two datasets, ACP135 and ACP99. The model achieved accuracy rates of 0.935 and 0.984, respectively—improving over existing methods by +2.6% (ACP135) and +5.8% (ACP99).
Conclusions
ACP-EPC is a powerful and interpretable framework for ACP prediction. By combining advanced structural/physicochemical features with cross-attention fusion, it outperforms prior approaches. The web interface visualizes per-residue attention heatmaps for each prediction.
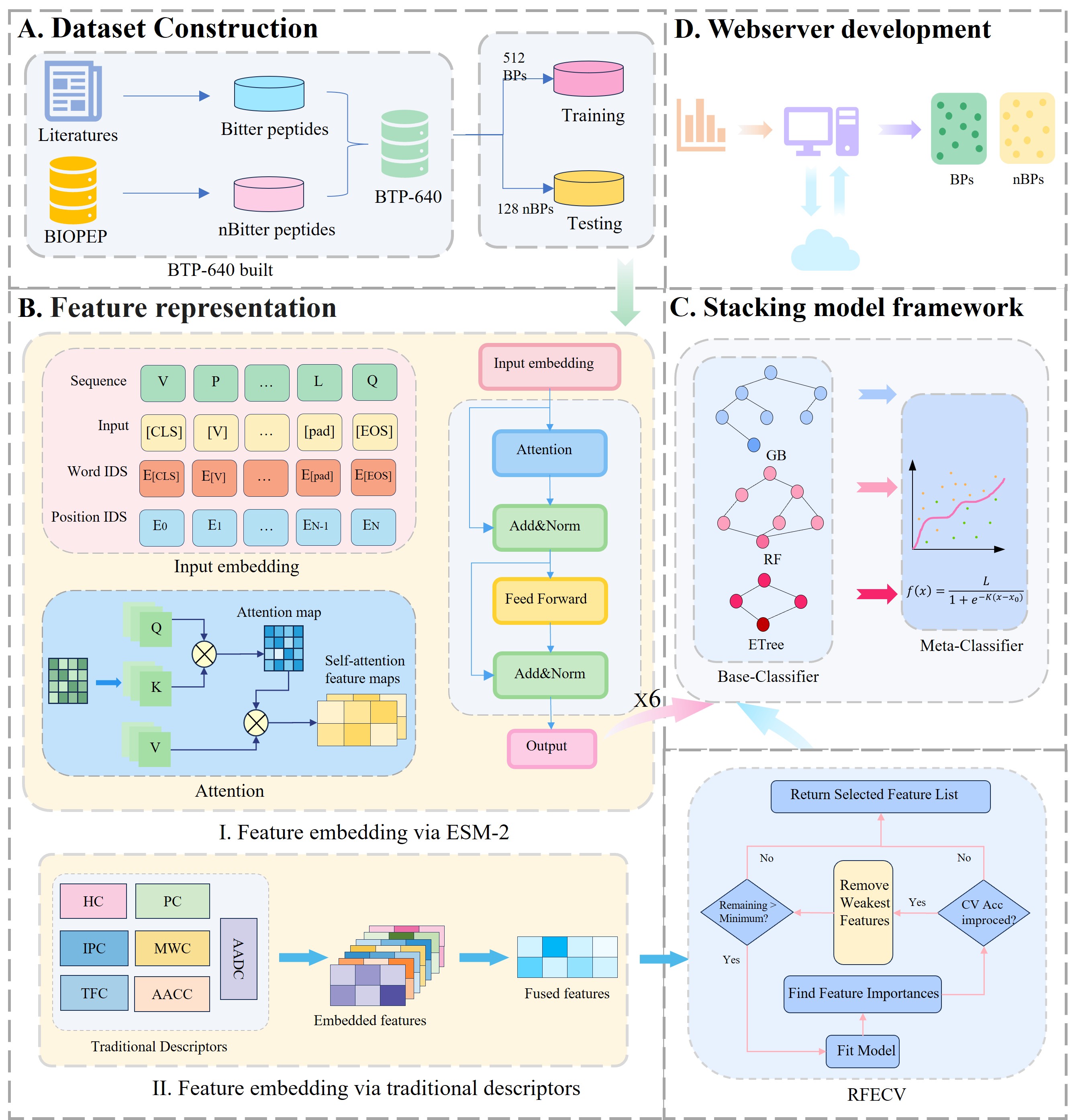
Bitter Peptides iBitter-GRE: A Novel Stacked Bitter Peptide Predictor with ESM-2 and Multi-View Features
Stacked learner • ESM-2 + traditional physicochemical descriptors • SHAP analysis
We proposed a stacked learner that combines ESM-2 representations and traditional physicochemical descriptors to identify bitter peptides. SHAP analyses quantified feature contributions and expert validation confirmed the model’s ability to capture key biochemical characteristics. Benchmarking showed superior accuracy over existing methods, and we provide a public web service for researchers.