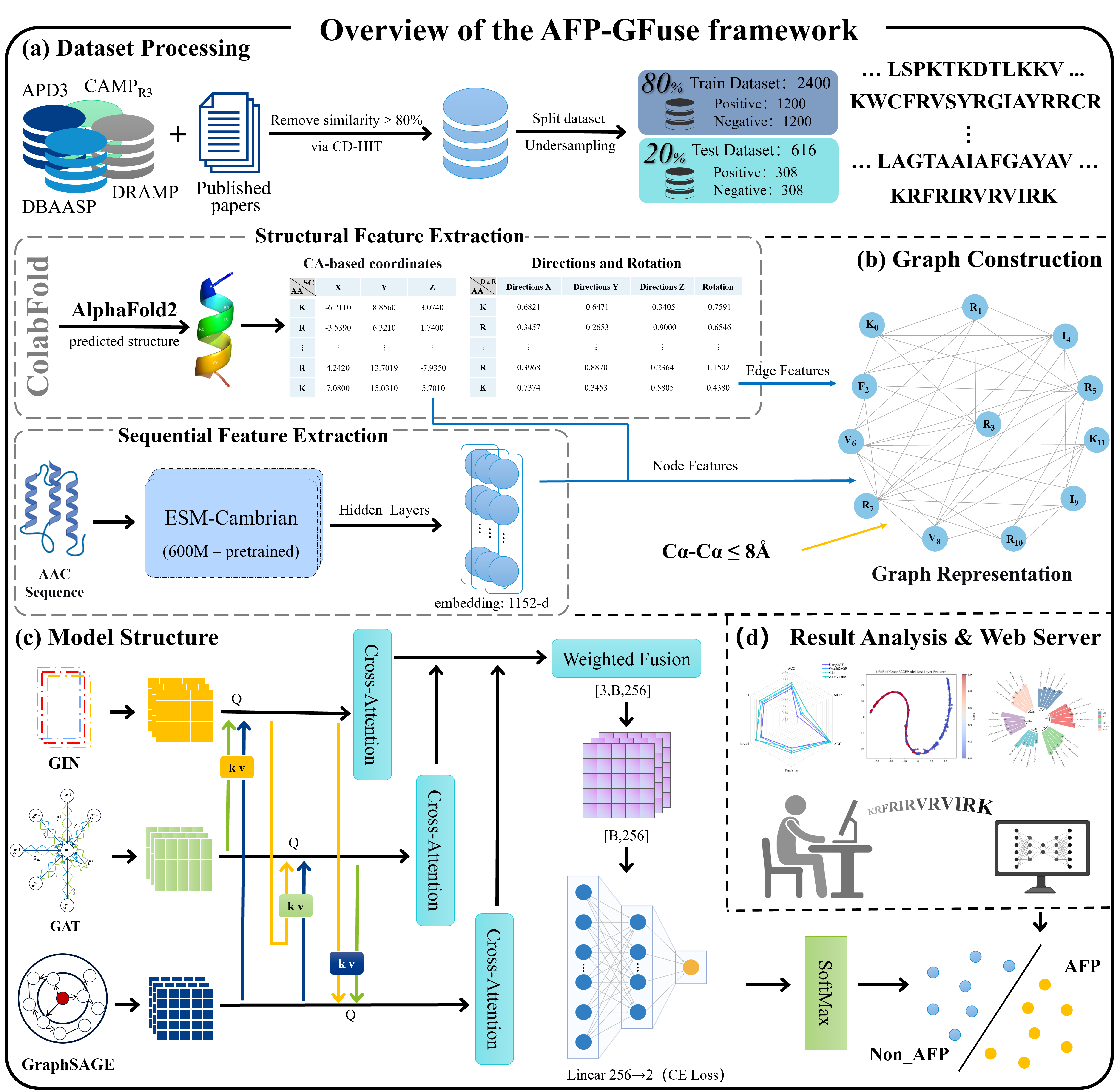
About AFP-GFuse
Antifungal peptides (AFPs) are endogenous defense agents that suppress fungal pathogens and help block invasion. Because their modes of action can circumvent common resistance pathways, AFPs often combine broad activity with favorable safety. However, traditional laboratory identification is slow and costly, spurring AI-based screening. Most existing computational tools rely on literature-compiled datasets and underrepresent AFP biology—e.g., they overlook 3D/spatial descriptors—and single graph neural networks (GNNs) tend to bias certain feature types. To close these gaps, we curated a current, comprehensive dataset and introduce AFP-GFuse, a deep model that unifies sequence signals with predicted structural information and couples three complementary GNN backbones. A hierarchical cross-attention module dynamically aligns and fuses multi-graph representations. In benchmarks, AFP-GFuse outperforms state-of-the-art baselines for AFP prediction (accuracy 0.9140). Ablation studies confirm the value of structural features, and comparisons against individual GNNs show that cross-attention fusion mitigates representational bias and boosts overall performance.
Short tagline:
“AFP-GFuse unifies sequence + 3D cues with hierarchical cross-attention over complementary GNNs, achieving 0.9140 accuracy and surpassing current SOTA for AFP prediction.”