Input Description
The input must be in FASTA format. Please enter your sequence here or select a file.
2.The system will default to the selected file, ensuring that the input file meets the format of the example.
Online Tool
Ensemble Predictor
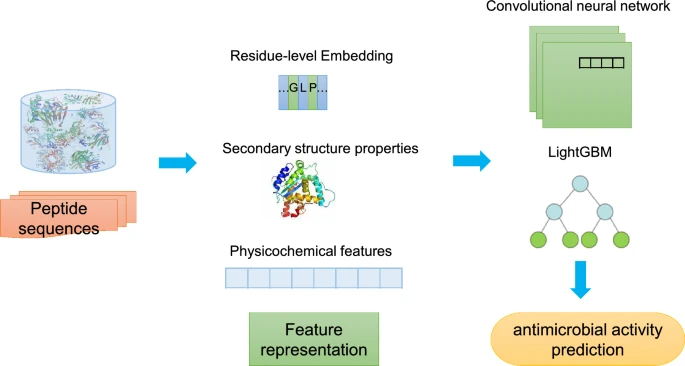
The emergence of antibiotic-resistant microbes raises a pressing demand for novel alternative treatments. One promising alternative is the antimicrobial peptides (AMPs), a class of innate immunity mediators within the therapeutic peptide realm. AMPs offer salient advantages such as high specificity, cost-effective synthesis, and reduced toxicity. Although some computational methodologies have been proposed to identify potential AMPs with the rapid development of artificial intelligence techniques, there is still ample room to improve their performance. This study proposes a predictive framework which ensembles deep learning and statistical learning methods to screen peptides with antimicrobial activity. We integrate multiple LightGBM classifiers and convolution neural networks which leverages various predicted sequential, structural and physicochemical properties from their residue sequences extracted by diverse machine learning paradigms. Comparative experiments exhibit that our method outperforms other state-of-the-art approaches on an independent test dataset, in terms of representative capability measures. Besides, we analyse the discrimination quality under different varieties of attribute information and it reveals that combination of multiple features could improve prediction. In addition, a case study is carried out to illustrate the exemplary favorable identification effect. We establish a web application at http://amp.denglab.org to provide convenient usage of our proposal and make the predictive framework, source code, and datasets publicly accessible at https://github.com/researchprotein/amp.
© BioAi-Lab All Rights Reserved