Input Description
The input must be in FASTA format. Please enter your sequence here or select a file.
2.The system will default to the selected file, ensuring that the input file meets the format of the example.
Online Tool
Stack-AVP Predictor
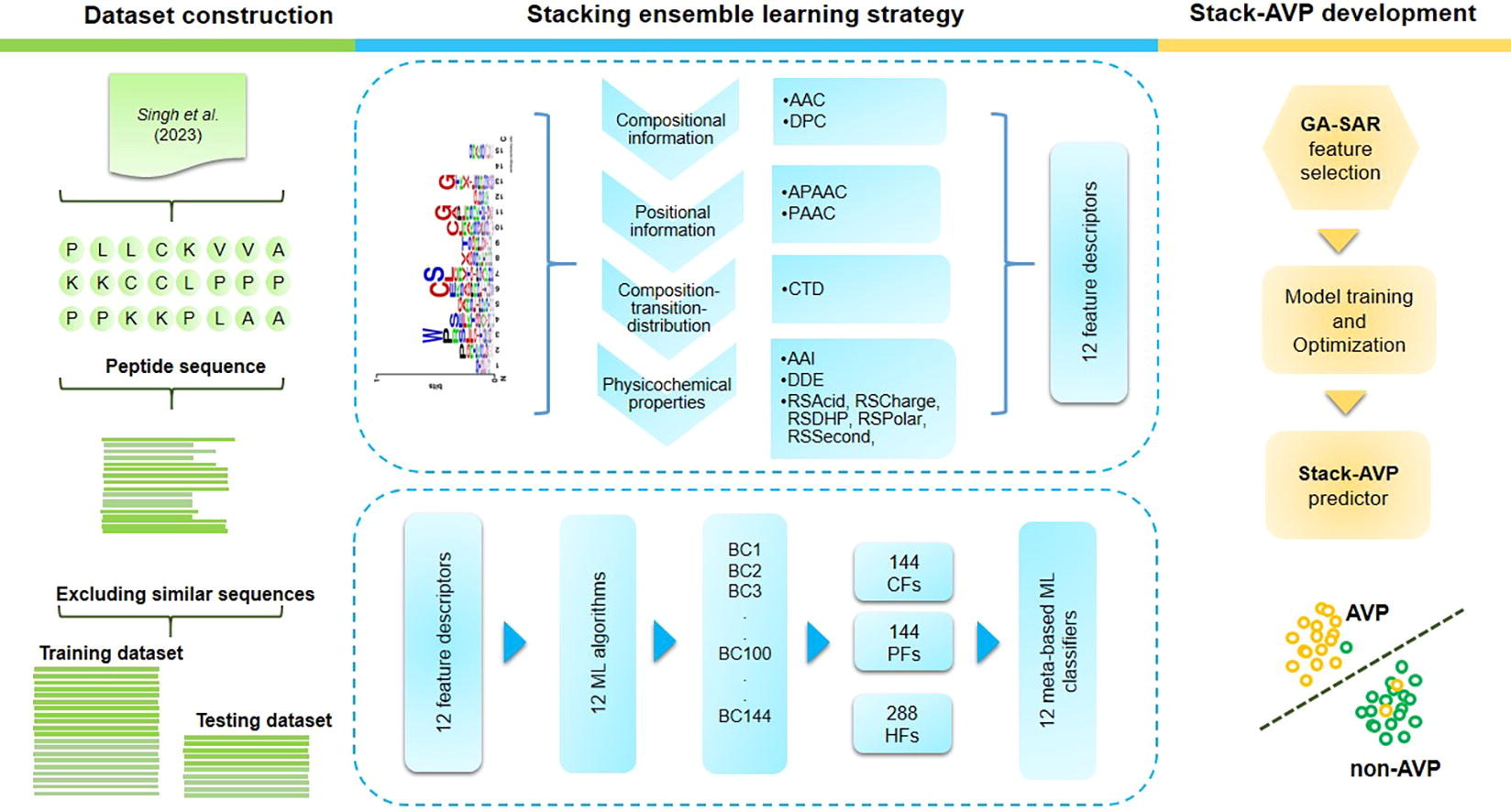
AVPs, or antiviral peptides, are short chains of amino acids capable of inhibiting viral replication, preventing viral entry, or disrupting viral membranes. They represent a promising area of research for developing new antiviral therapies due to their potential to target a broad spectrum of viruses, incorporating those resistant to traditional antiviral drugs. However, traditional experimental methods for identifying AVPs are often costly and labour-intensive. Thus far, multiple computational methods have been introduced for the in silico identification of AVPs, but these methods still have certain shortcomings. In this study, we propose a novel stacked ensemble learning framework, termed Stack-AVP, for fast and accurate AVP identification. In Stack-AVP, we investigated heterogeneous prediction models, which were trained with 12 commonly used machine learning algorithms coupled with a wide range of multiple feature encoding schemes. Subsequently, these prediction models were adopted to generate multi-view features providing class information and probability information. Finally, we applied our feature selection method to determine the best feature subset for the construction of the final stacked model. Comparative assessments on the independent test dataset revealed that Stack-AVP surpassed the performance of current state-of-the-art methods, with an accuracy of 0.930, MCC of 0.860, and AUC of 0.975. Furthermore, it was found that our multi-view features exhibited a crucial mechanism to improve the prediction performance of AVPs. To facilitate experimental scientists in performing high-throughput identification of AVPs, the prediction sever Stack-AVP is publicly accessible at https://pmlabqsar.pythonanywhere.com/Stack-AVP.
© BioAi-Lab. All Rights Reserved.