Input Description
The input must be in FASTA format. Please enter your sequence here or select a file.
2.The system will default to the selected file, ensuring that the input file meets the format of the example.
Online Tool
Hyb_SEnc Predictor
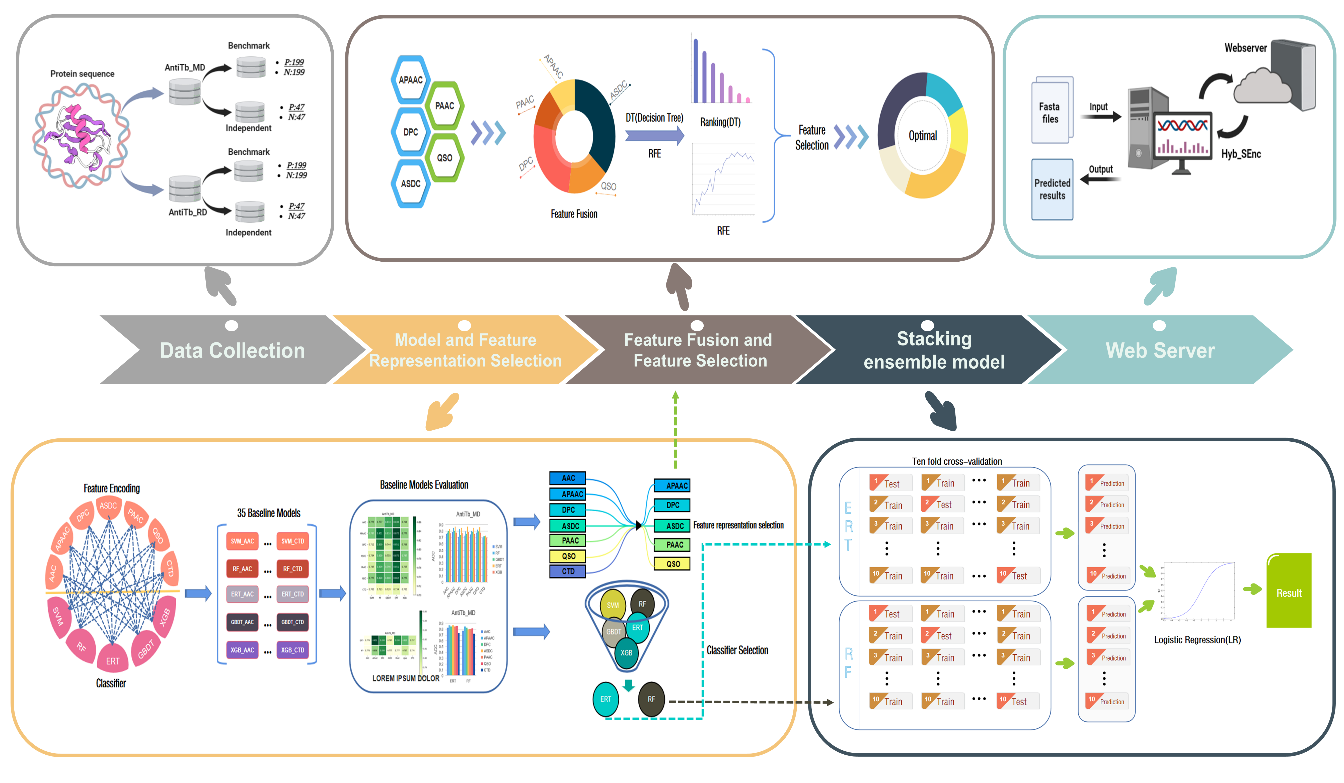
Tuberculosis has plagued mankind since ancient times, and the struggle between humans and tuberculosis continues. Mycobacterium tuberculosis is the leading cause of tuberculosis, infecting nearly one-third of the world's population. The rise of peptide drugs has created a new direction in the treatment of tuberculosis. Therefore, for the treatment of tuberculosis, the prediction of anti-tuberculosis peptides is crucial. This paper proposes an anti-tuberculosis peptide prediction method based on hybrid features and stacked ensemble learning. First, a random forest (RF) and extremely randomized tree (ERT) are selected as first-level learning of stacked ensembles. Then, the five best-performing feature encoding methods are selected to obtain the hybrid feature vector, and then the decision tree and recursive feature elimination (DT-RFE) are used to refine the hybrid feature vector. After selection, the optimal feature subset is used as the input of the stacked ensemble model. At the same time, logistic regression (LR) is used as a stacked ensemble secondary learner to build the final stacked ensemble model Hyb_SEnc. The prediction accuracy of Hyb_SEnc achieved 94.68% and 95.74% on the independent test sets of AntiTb_MD and AntiTb_RD, respectively.
© BioAi-Lab All Rights Reserved