Input Description
The input must be in FASTA format. Please enter your sequence here or select a file.
2.The system will default to the selected file, ensuring that the input file meets the format of the example.
Online Tool
iBitter-GRE Predictor
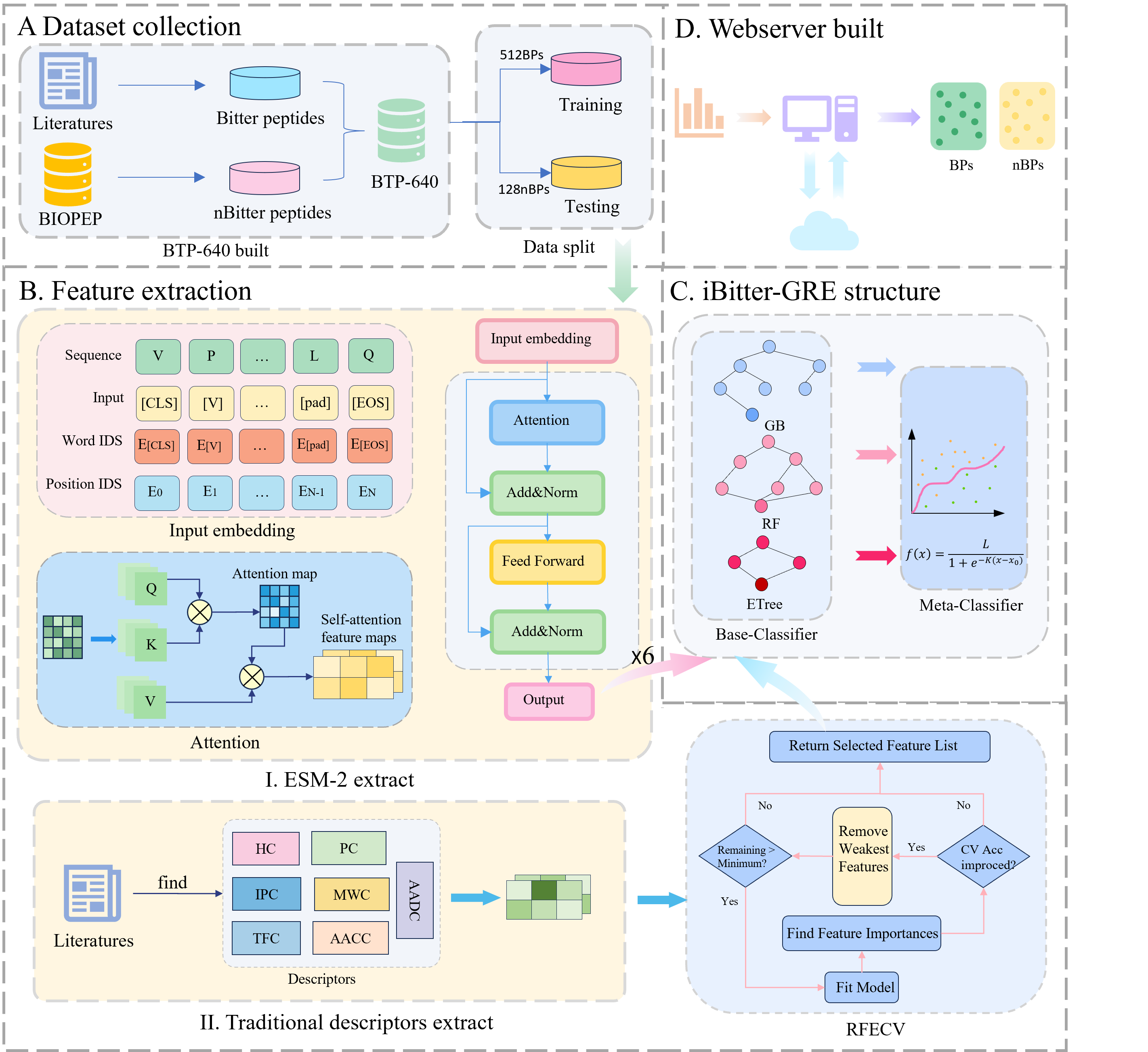
Accurate identification of bitter peptides is essential for research. Although models using sequence information have evolved in the context of bitter peptides, there is still room for improvement in their predictive performance. In the present study, we introduced a novel predictive tool, iBitter-GRE, designed to improve the accuracy of bitter peptide identification. Our model uses ESM-2 and traditional descriptors capture the physical and biochemical properties of bitter peptides for feature extraction. To expand the model’s learning capabilities, we adopted a stacking approach to integrate multiple learners. Feature contributions were analyzed using SHAP values. Validation by domain experts confirmed that our model effectively identifies the key biochemical characteristics of bitter peptides. Benchmark experiments showed that iBitter-GRE achieves higher accuracy than existing methods. To assist the researchers, we created a web server accessible at http://www.bioai-lab.com/iBitter-GRE. We believe that iBitter-GRE is a valuable tool for the discovery and identification of bitter peptides.
© BioAi-Lab All Rights Reserved