 |
DeepMC-iNABPA deep learning-based multi-class classifer for nucleic acid binding protein |
 |
DeepMC-iNABPA deep learning-based multi-class classifer for nucleic acid binding protein |
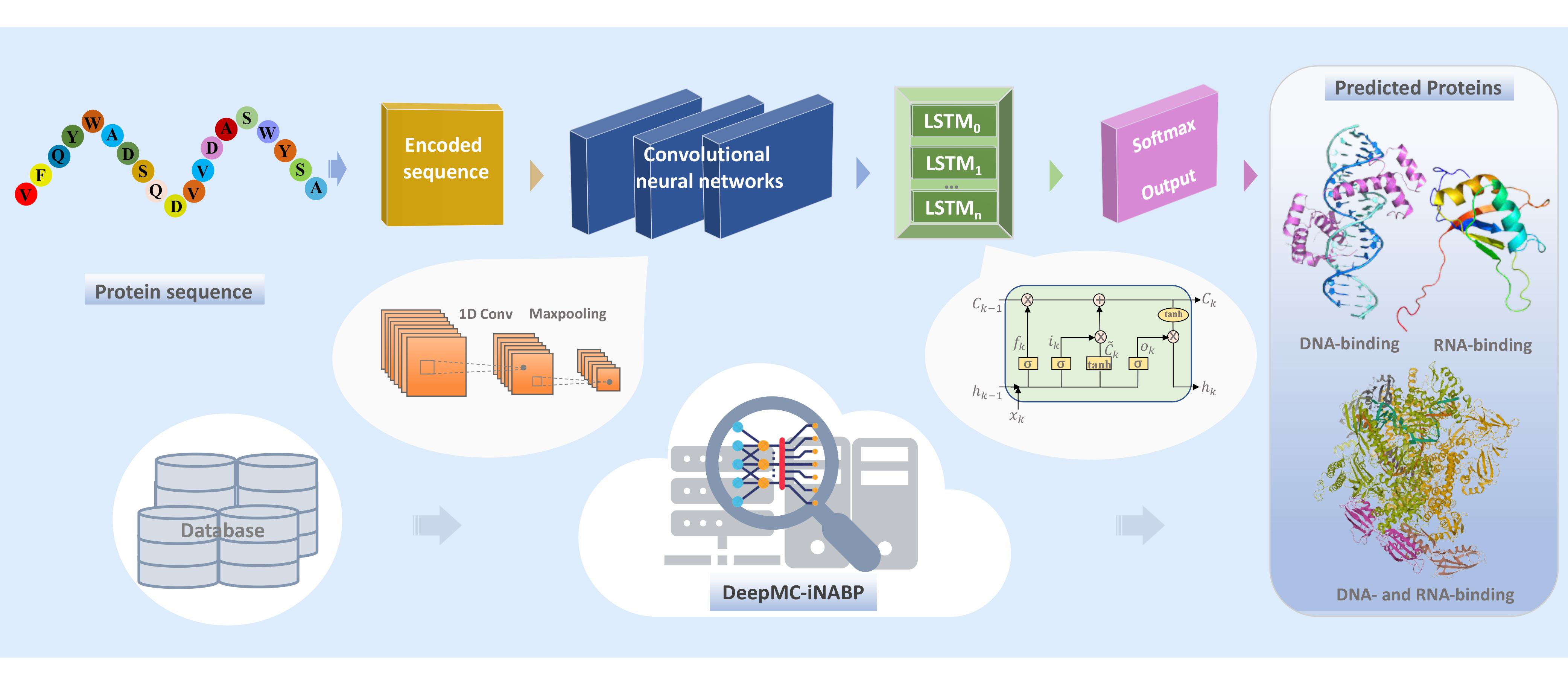
Nucleic acid binding proteins (NABPs) including DNA-binding proteins (DBPs) and RNA-binding proteins (RBPs), play vital roles in gene expression. Accurate identification of them is crucial important. In previous studies, there are two existing challenges: one is the problem of ignoring DNA- and RNA-binding proteins (DRBPs), the other is cross-predicting problem referring to DBP predictors predict DBPs as RBPs, and vice versa. In this study, we proposed a computational predictor, called DeepMC-iNABP, aiming at solve these difficulties by utilizing multi-class classification strategy and deep learning approaches. DBPs, RBPs, DRBPs and non-NABPs as separate classes data were used for training the DeepMC-iNABP model. The results on test data collected in this study and two independent test datasets showed that DeepMC-iNABP has strong advantage in identifying the DRBPs and has ability of alleviating the cross-prediction problem to a certain extent.
| Download | Description | |
|---|---|---|
| Data (in this study) | DBPdata RBPdata DRBPdata notNABPdata | DNA-binding protein Data RNA-binding protein Data DNA- and RNA-binding protein Data not Nucleic Acid-binding protein Data |
| Independent Data | TEST474data DRBP206data | TEST474 Independent Dataset DRBP206 Independent Dataset |
| Code Resources | DeepMC-iNABP code | Code Resources for DeepMC-iNABP model |
| Feifei Cui#, Shuang Li#, Zilong Zhang, Miaomiao Sui, Chen Cao, Abd El-Latif Hesham, Quan Zou*. DeepMC-iNABP: Deep learning for multiclass identification and classification of nucleic acid-binding proteins. Computational and Structural Biotechnology Journal. 2022, 20: 2020-2028. |
| Feifei Cui | feifecui0910@gmail.com | ||
| Quan Zou | zouquan@nclab.net | ||