AVP-HNCL
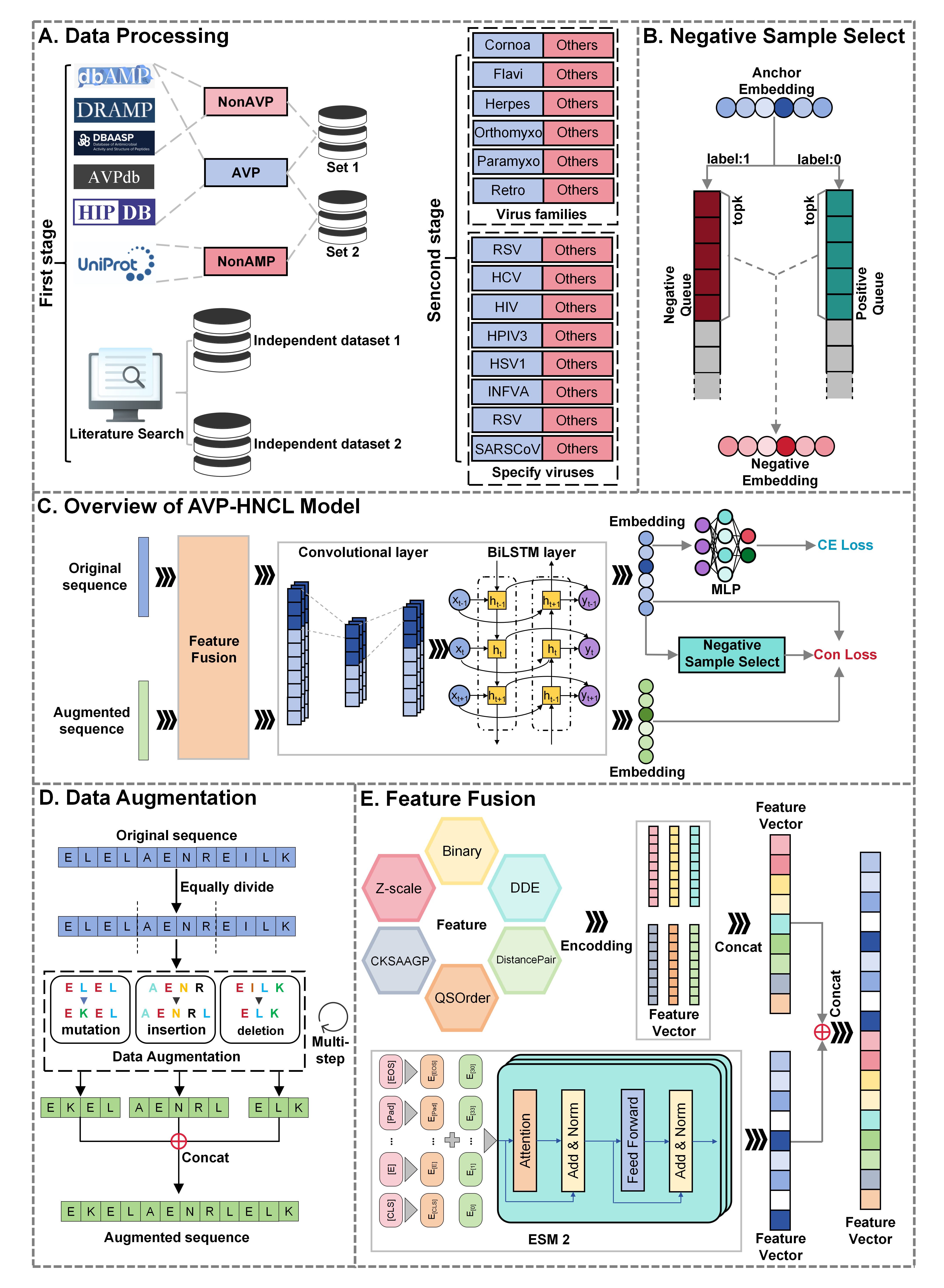
Antiviral peptides (AVPs) hold significant potential in public health by effectively inhibiting a variety of viruses. However, existing research primarily relies on traditional models to predict their activity, which are limited by feature extraction capabilities and model generalization. To address these limitations, we propose a two-stage predictive framework that integrates the ESM2 model, data augmentation, feature fusion, and contrastive learning techniques. Utilizing an innovative top‑k queue-based contrastive learning strategy, this framework significantly enhances the accuracy and generalization performance in identifying AVPs and their subclasses. The model demonstrates excellent performance on independent datasets, showcasing the strong application potential of this method in antiviral peptide research.
Use AVP-HNCL to predict antiviral peptide activity:
Resources
Access the code and data for AVP-HNCL on our GitHub repository.
GitHub RepositoryAbout Us
We are a laboratory dedicated to bioinformatics research, with the goal of using computational methods to analyze biological sequence data to support life science research.
Contact Us
Please address your comments, questions, and suggestions to:
Feifei Cui: feifeicui@hainanu.edu.cn
Zilong Zhang: zhangzilong@hainanu.edu.cn
To learn more, please visit our laboratory's official website:
Visit Official Website