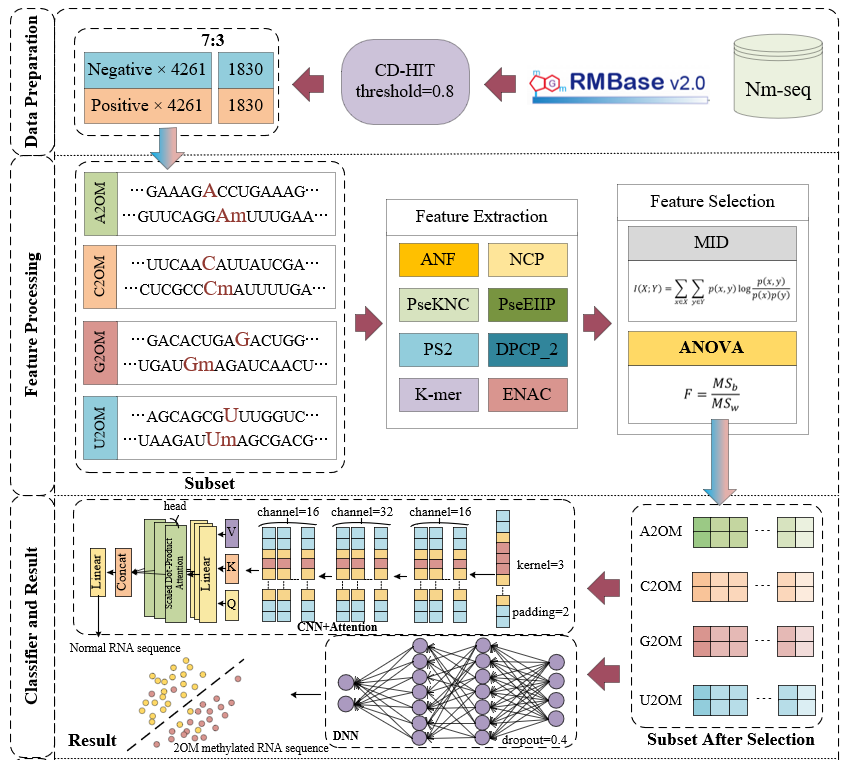
DeepR2OM
Main Figure
RNA Site Lab
1.Enter your sequence here, or select a file. You can choose either a TXT or FASTA file.
2.The system will default to the selected file, ensuring that the input file meets the format of the example.
RNA sequence length must be equal to 41

Main Figure
RNA Site Lab
© RNASite. All Rights Reserved.