1. How to use Database ?
The method of using databases :
The RNA site contained in the database are already provided in the drop-down menu. Each locus includes at least one dataset, the corresponding identification tool, the description of the dataset, the link to the original text, and the dataset download.
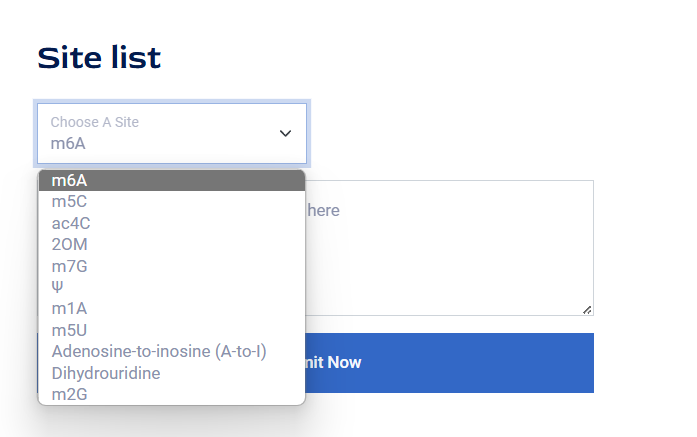
1.Select a site you wish to search for from the drop-down menu as shown in the figure.
2.Click the Submit button.
3.You will obtain the dataset information related to this RNA Site in the database on a new page.
2. How to use Servers ?

The method of using RNA site recognition tools:
The RNA site contained in the database are already provided in the drop-down menu. Each locus includes at least one dataset, the corresponding identification tool, the description of the dataset, the link to the original text, and the dataset download.
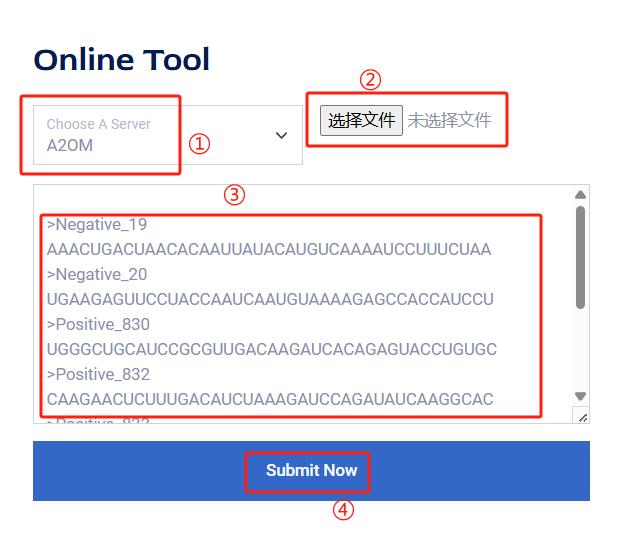
1.Select a server to use from the navi bar; for example, let's take the 2OM server.
2.When on the server page, you will see a dialog box as shown in the figure:
① Choose a server you want to use for the identification task.
② You can select a .txt or .fasta file you want to use for the identification task.
③ If the number of sequences is small, you can directly copy them into the dialog box.
④ Click the Submit button to obtain the results.
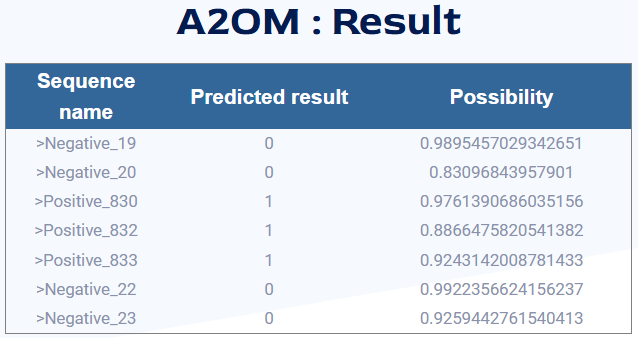
3.On the results page:
The first column is the ID.
The second column is the classification status, with 1 indicating positive and 0 indicating negative.
The third column (if any) is the probability of classification.
4.You can click the Download button on the results page to save your predicted results to your local storage.